Keywords
SARS-CoV-2, adolescents, COVID-19 waves, Uganda, seropositivity, longitudinal study
COVID-19 remains a significant global public health concern, contributing to hospitalization and Intensive Care Unit (ICU) admissions. However, SARS-CoV-2 exposure in adolescents remains poorly understood. The study aimed to analyse temporal trends in SARS-CoV-2 seropositivity in Ugandan adolescents using pre-pandemic (2014–2016) and pandemic-era samples (2020–2022).
We analysed stored blood samples from a subset of participants enrolled in the Entebbe Mother and Baby Study (EMaBS), a long-standing Ugandan birth cohort established in 2003 as a randomized trial (ISRCTN32849447). Samples were drawn from three studies: pre-pandemic samples (2014–2016) collected during a blood pressure study; pandemic-era samples collected during the POPVAC C trial (2020–2021), which evaluated the impact of BCG revaccination on vaccine responses; and samples from the CoHost study (2021–2022), a 12-month longitudinal follow-up assessing SARS-CoV-2 infection and exposure. Blood samples were collected at multiple study timepoints. SARS-CoV-2 IgG spike antibodies were measured using an in-house enzyme-linked immunosorbent assay (ELISA).
None of the 47 participants were seropositive for SARS-CoV-2 in pre-pandemic samples (2014–2016). In pandemic-era samples (2020–2022), seropositivity increased over time. At POPVAC C screening, 2 (4%) participants were seropositive, increasing to 3 (6%) at week 4, 5 (11%) at week 8, and 36 (77%) at week 52, which also served as the CoHost baseline. At the CoHost 6-month timepoint, 40 (85%) of participants were seropositive, increasing to 45 (96%) at 12 months timepoint. SARS-CoV-2 seropositivity progressively increased, with a marked rise during the Delta wave (May‒July 2021) and peaking in the Omicron wave (November 2021 - February 2022).
Our findings indicate increased SARS-CoV-2 exposure among adolescents during later pandemic waves, likely driven by the emergence of highly transmissible variants and changes in public health measures. These preliminary findings highlight the need for age-specific mitigation strategies.
COVID-19 is a serious illness that has affected people around the world, but we know less about how it has spread among adolescents, especially in African settings like Uganda. In this study, we looked at blood samples from Ugandan teenagers who are part of a long-running health study that began when they were babies.
We used stored blood samples taken before the pandemic (between 2014 and 2016) and during the pandemic (from 2020 to 2022) to see how many adolescents had signs of past COVID-19 infection. To do this, we tested their blood for antibodies—proteins made by the immune system in response to the virus.
We found that none of the adolescents had antibodies before the pandemic. During the pandemic, more and more teenagers showed signs of past infection as time went on. By the end of the study in 2022, nearly all of them had been exposed to the virus. The biggest increases happened during the Delta and Omicron waves of COVID-19, when the virus was spreading quickly.
This study shows that many adolescents in Uganda were infected during the later waves of the pandemic. It suggests that public health efforts—like targeted vaccination or school safety measures—should take age into account when planning how to protect communities in future outbreaks.
SARS-CoV-2, adolescents, COVID-19 waves, Uganda, seropositivity, longitudinal study
COVID-19 remains a significant global public health concern, contributing to hospitalization and ICU admissions1. SARS-CoV-2, the causative agent of COVID-19 was first reported in December 2019 in Wuhan, China2. On January 30, 2020, the World Health Organization (WHO) declared the outbreak a Public Health Emergency of International Concern (PHEIC), a status that remained in effect until May 5, 20233. The virus rapidly spread worldwide leading WHO to classify COVID-19 as a pandemic on March 11, 20204. However, the removal of the PHEIC status did not signify eradication, and public health efforts remain essential to manage the long-term impact of the virus.
The COVID-19 pandemic in Africa unfolded in four distinct wave periods5. The first wave was driven by the prototype SARS-CoV-2 strain while the subsequent waves were characterised by the emergence of specific variants, such as Alpha, Beta, Eta (second wave); Delta (third wave); and Omicron (fourth wave). These waves exhibited regional variation, with some variants predominating in specific areas. For example the Eta variant, first detected in Nigeria in November 2020 was prevalent in West Africa during the second wave but remained rare in Eastern and Southern Africa5.
Uganda reported its first COVID-19 case on March 21, 2020, and subsequently experienced three distinct waves of infection6. The first wave (December 2020‒January 2021) was dominated by A.23.1 lineage which was initially detected in Uganda7. The second wave (May‒July 2021) was driven by the Delta variant, while the third wave (November 2021 - February 2022) involved both Delta and the emerging Omicron variant6.
In addition to different transmission trends, these waves exhibited distinct clinical presentations and outcome8. The Delta variant was associated with higher disease severity and hospitalization rates, whereas Omicron was linked to milder disease presentations in most individuals8. In a prospective longitudinal observational study, symptom profiles differed between Delta and Omicron infections: loss of smell was significantly less common in Omicron cases than in delta cases (16·7% vs 52·7%, OR 0·17; 95% CI 0·16–0·19, p<0·001)9. Conversely, sore throat was more frequently reported during Omicron infections than Delta (70·5% vs 60·8%, OR 1·55, 95% CI 1·43–1·69, p<0·001). Furthermore, there was a lower rate of hospital admission during Omicron than during Delta (1·9% vs 2·6%, OR 0·75; 95% CI 0·57–0·98, p=0·03)9.
However, transmission dynamics also shifted over time, particularly across age groups. While early waves were associated with low hospitalisation rates in children, the Omicron wave led to a rise in paediatric hospital admissions10.
Despite these insights, adolescent exposure to SARS-CoV-2 in Africa remains poorly understood. Questions remain regarding their exposure and how pandemic induced socioeconomic disruptions may have increased their vulnerability5. To address these gaps, we aimed to analyse temporal trends in SARS-CoV-2 seropositivity among adolescents in a longitudinal pilot study nested in a birth cohort before and during distinct COVID-19 waves in Uganda.
From the inception of the study, there was active involvement of participants and members of the public. A consultation committee was established, comprising parents, adolescent participants, and respected community representatives. This committee met regularly—both prior to study initiation and throughout the study period—to advise on study procedures and documentation. The goal of this engagement was to ensure that participant and community perspectives were meaningfully integrated throughout the project and to support the research team in aligning study activities with the local context. Study findings will be disseminated through peer-reviewed publications, conference presentations, and direct feedback to participants and their families.
We utilized a subset of samples from a longitudinal study nested within the Entebbe Mother and Baby Study (EMaBS) birth cohort, based in Entebbe municipality, Uganda. Established in 2003 as a randomized trial (ISRCTN32849447), EMaBS was originally designed to assess whether treating worm infections during pregnancy and early childhood influences immune responses to vaccines, the incidence of infectious diseases, and allergy-related outcomes11.
The nested longitudinal study, CoHost, titled life-course determinants of susceptibility and response to SARS-CoV-2 infection in EMaBS participants and their role in transmission and control, actively followed up participants for 12 months (August 2021–December 2022) to monitor SARS-CoV-2 infection and exposure. Blood samples were collected at three main study time points—month 0 (August 30 to November 22, 2021), month 6 (February 25 to May 20, 2022), and month 12 (August 25 to December 14, 2022), alongside additional data obtained via questionnaires and diary cards. In accordance with the Ministry of Health directive, participants were adminstered COVID-19 vaccination between month 6 and month 12 timepoints.
Prior to CoHost, a subset of EMaBS participants (who also later took part in CoHost) were enrolled in POPVAC C (ISRCTN10482904), an open-label, randomized controlled trial conducted between August 2020 and October 2021. This trial assessed the effect of BCG revaccination on the immunogenicity of unrelated vaccines12. Blood samples were collected and stored at screening (August 31, to October 12, 2020), week 4 (September 28, to November 13, 2020), week 8 (October 6, to December 7, 2020), and week 52 (August 30, to October 21, 2021).
As part of earlier work in the EMaBS birth cohort, stored samples from a blood pressure (BP) study were collected when participants were 10–11 years (between May 2, 2014 and June 1, 2016); these were utilised as pre-pandemic samples in this analysis13.
Previously stored demographic and clinical data, along with bio banked samples, were used for this study. In the CoHost study, participants were administered a questionnaire capturing demographic information, medical history, SARS-CoV-2 exposure, and recent illness history, recorded using the research electronic data capture (REDCap) application.
As described above, stored samples collected at three time points in the CoHost study, four timepoints in the POPVAC C trial and one time point in the earlier EMaBS BP study were used for this analysis.
This study was conducted as a pilot within the CoHost study to explore optimal timepoints for assessing SARS-CoV-2 seropositivity. All these blood samples were drawn from peripheral veins, and serum samples stored at –80 °C at MRC/UVRI and LSHTM Uganda Research Unit in Entebbe, Uganda.
Specific IgG antibodies to the spike antigen (S-antigen) of the COVID-19 virus were measured using an in-house IgG ELISA. Spike protein antigen consists of a pre-fusion S ectodomain residues 1–1138 with proline substitutions at amino acid positions 986 and 987, a GGGG substitution at the furin cleavage site (amino acids 682–685) and an N terminal T4 trimerisation domain. Medium binding 96 well plates were coated overnight with 50μl per well of S-antigen (kindly provided by Katie J Doores’ laboratory, Department of Infectious Diseases, King’s College London, UK) at a coating concentration of 3μg/ml. The plates were washed with phosphate-buffered saline (PBS 1X)-Tween 20 (0.05%) (Sigma-Aldrich, UK) solution and blocked with 200μl of 3% skimmed milk (Sigma-Aldrich, UK) diluted in PBS-Tween 20 (Sigma-Aldrich, UK) for 1 hour at room temperature (RT). The plates were rewashed and incubated for 1 hour at 37 oC with 50μl of test plasma, positive and negative control samples (diluted 1/100 with 3% skimmed milk in PBS-Tween 20). Positive controls were derived from a pooled sample generated from sera of known COVID-19 samples, kindly provided by Jennifer Serwanga, MRC/ UVRI and LSHTM Uganda Research Unit. Following the test, positive and negative control sample incubation, plates were washed and detection antibody conjugated to goat anti-human IgG-horseradish peroxidase conjugate (Insight Biotechnology, UK), diluted 1/3000 in assay buffer was added. The plates were incubated for 1 hour at RT, rewashed, and developed by addition of 50μl of 3,3',5,5'-tetramethylbenzidine (Sigma-Aldrich, UK) and reactions stopped after 10 minutes with 100μl of 2M hydrochloric acid (Fisher Scientific, UK). Optical density (OD) values were measured at 450nm on a 96-well plate ELISA reader (BioTek ELx808, USA). To minimise batch effects, all samples were randomised across all plates and done in a single run to ensure uniform experimental conditions and reduce potential variability which could arise from batch-to-batch differences.
All statistical analyses were conducted using Stata version 18.5 (StataCorp, College Station, TX, USA). A descriptive analysis of the participants' characteristics was conducted. Descriptive statistics were performed to summarize the distribution of optical density measurements of spike protein IgG seropositivity across different time points. Boxplots were used to visually depict the median, interquartile range (IQR), and outliers for each time point. The epidemiological curve was plotted based on WHO COVID-19 data for Uganda obtained from https://data.who.int/dashboards/covid19/data with the periods of time we collected samples of participants highlighted.
Ethical approvals for the studies from which samples were used were obtained from the Uganda Virus Research Institute Research and Ethics Committee (UVRI REC) (Reference no. - GC/127/35) dated June 23, 2021, the Uganda National Council for Science and Technology (UNCST) (Reference no. - MV 625) dated September 2, 2020, the Uganda National Drug Authority (for the POPVAC C clinical trial) (Reference no. - CTA0094) dated May 28, 2019, and the London School of Hygiene & Tropical Medicine (LSHTM) Research Ethics Committee (Reference no. - 8811) dated July 12, 2021. Written informed consent was obtained for participants aged 18 years and above, parental consent was obtained for participants aged 17 years and younger, and assent was obtained for participants aged 8–17 years, as per local regulatory requirements. All study activities were conducted in accordance with the local ethical guidelines and adhered to the principles outlined in the Declaration of Helsinki on ethical principles for medical research involving human participants14.
Figure 1 illustrates the daily reported SARS-CoV-2 cases in Uganda from April 2020 and December 2022, highlighting three distinct pandemic waves. The first wave (A.23.1 variant) occurred between December 2020 and January 2021. The second wave (Delta variant), peaked between May and July 2021, while the third wave (Omicron variant) emerged between December 2021 and February 2022. The timeline also marks nationwide public health interventions, including two lockdowns and mandatory school closures, alongside COVID-19 vaccination timepoints and study-related sampling points for pandemic era samples from the POPVAC C and CoHost studies. POPVAC C samples were collected at week 0, 4 and 8 during the first wave (August 2020-December 2020), while week 52 sampling which coincided with the CoHost baseline, was conducted between the second and third waves (August 2021 - November 2021). CoHost follow up samples at month 6 and 12, were collected after the third wave between February -May 2022 and August-December 2022, respectively.
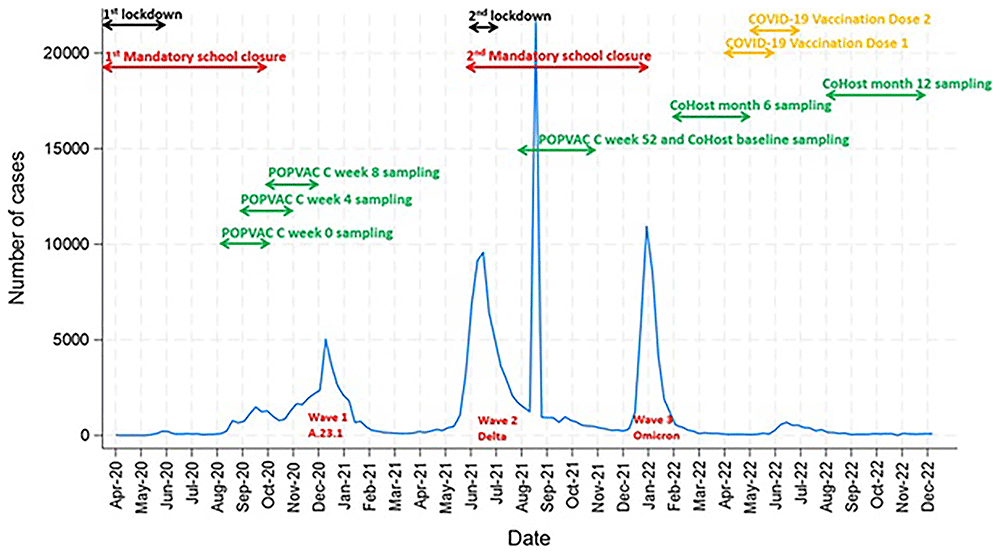
Data source: https://data.who.int/dashboards/covid19/data *The apparent spike in cases between August and September 2021 reflects data harmonization by the Ministry of Health, reconciling discrepancies between laboratory-confirmed cases and surveillance reports15.
This pilot analysis included stored samples retrieved for 47 participants in the birth cohort who were enrolled in all three studies. The mean age of the participants providing pandemic-era samples was 16.6 years (SD = 0.8, range 15 – 17), and 59.6% were male. All participants were confirmed HIV-negative.
During the CoHost study follow up period (August 2021-November 2022), 8 (17%) participants reported experiencing COVID-19-like illness and underwent PCR testing; however, only one tested positive. None of the participants had ever received a COVID-19 vaccination at the time of enrolment. During follow up, 21 (44.7%) participants received 2 doses of COVID-19 vaccination between the CoHost month 6 and month 12 timepoints.
No participants showed seropositivity for SARS-CoV-2 in pre-pandemic samples (2014–2016). In pandemic-era samples (2020–2022), seropositivity increased over time. At POPVAC C screening, 2 (4%) participants were seropositive, increasing to 3 (6%) at week 4, 5 (11%) at week 8, and 36 (77%) at week 52, which also served as the CoHost baseline. At the CoHost 6-month timepoint, 40 (85%) participants were seropositive, rising to 45 (96%) at 12 months timepoint.
Mean optical density (OD) values increased progressively across successive timepoints, reflecting increasing seropositivity (Figure 2). A significant rise in seropositivity was observed between POPVAC C week 8 and week 52. By the final CoHost timepoint, nearly all participants had seroconverted. 3 of 21 participants vaccinated between Month 6 and 12 seroconverted post-vaccination. In Figure 2, the red line represents seropositivity threshold cut off of 0.454 used16. Notably, this increase in seropositivity closely coincided with the subsequent COVID-19 waves in Uganda (Figure 2).
This preliminary analysis evaluated temporal trends in SARS-CoV-2 seropositivity among adolescents in a longitudinal pilot study nested in a birth cohort, spanning different COVID-19 waves in Uganda. Our findings indicate a progressive increase in seropositivity over time, with a marked rise during the Delta wave and peaking in the Omicron wave. This suggests likely increased exposure to SARS-CoV-2 among adolescents in later phases of the pandemic. This may be due to the emergence of highly transmissible variants, such as the Delta and Omicron variants, or vaccine uptake in some of the participants or changes in public health measures, such as removal of lockdowns and reopening of schools. Furthermore, strict public health measures, including lockdowns and school closures, may have limited SARS-CoV-2 exposure in the early phases of the pandemic17. These trends highlight the dynamic nature of SARS-CoV-2 seropositivity in this cohort and underscore the role of community transmission in shaping it.
Our findings align with global seroprevalence trends observed among children and adolescents. A systematic review and meta-analysis reported an increase in global seroprevalence over time, rising from 7% during early waves to 37–56% during the Delta and Omicron waves18. In South Africa, adolescents exhibited higher seroprevalence rates than adults, particularly during Delta and Omicron waves19. Similarly, in Uganda, a significantly larger proportion of children were infected during the Delta and Omicron waves compared to the Alpha wave (71.6% versus 19.2%, p < 0.001)20. These findings underscore critical public health considerations, including the need for age-specific mitigation strategies, enhanced surveillance, vaccine prioritization, and adaptive school policies to minimize transmission and protect vulnerable populations.
A major strength of this study is its longitudinal design, which includes pre-pandemic samples and an extended follow-up period, enabling the assessment of seropositivity across multiple COVID-19 waves. However, some limitations should be considered. First, the study employed a convenience sample drawn from an established birth cohort, which may limit generalizability of findings to broader adolescent populations. Second, symptomatic SARS-CoV-2 infections may have been underreported, particularly during lockdown periods, as participants could have sought care elswhere. Third, the study focused exclusively on humoral immunity; we did not assess T-Cell responses, which are known to contribute substantially to long-term protection against SARS-CoV-221. Fourth, some participants received COVID-19 vaccination after the Omicron wave, complicating the interpretation of the spike-specific seropositivity, which may reflect natural exposure, vaccination or both. However, this may have unlikely affected the overall picture as most of the participants had already seroconverted before being vaccinated. Fifth, as this was a pilot study with a relatively small sample size, the findings should be considered preliminary and interpreted with caution. Further research with larger cohorts and more comprehensive immunological profiling is warranted to validate and extend these observations.
In summary, our findings suggest that SARS-CoV-2 seropositivity among adolescents increased during later pandemic waves, coinciding with the emergence of highly transmissible variants such as Delta and Omicron. Further research is needed to elucidate the interplay between SARS-CoV-2 variant evolution and immune responses over time. Additionally, studies assessing factors associated with SARS-CoV-2 susceptibility in adolescents are essential to inform future public health strategies, not only for COVID-19 but also for strengthening preparedness and response to future pandemics.
Data underlying this study is archived on the LSHTM Data Compass repository at https://doi.org/10.17037/DATA.0000467822. To ensure use complies with study ethics permissions, access to the data is subject to approval. However, these conditions are intended to be minimal and access will be provided without undue reservation.
We would like to thank Kate Naluwu for her support in conducting this preliminary laboratory work.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Register with NIHR Open Research
Already registered? Sign in
If you are a previous or current NIHR award holder, sign up for information about developments, publishing and publications from NIHR Open Research.
We'll keep you updated on any major new updates to NIHR Open Research
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)