Keywords
Bacteremia, Endocarditis, Diagnostic stewardship
International guidelines for S aureus bacteraemia often recommend screening echocardiography irrespective clinical factors that influence the likelihood of endocarditis, but this is not the case for enterococci or streptoccoci that are also high risk organism. Time to blood culture positivity (TTP) reflects bacterial load and has the potential to identify patients at higher risk of endocarditis.
We performed a retrospective analysis of clinical data that have been performed in a tertiary hospital in England. We investigate time to positivity (TTP) as a predictor variable in our S. aureus, enterococci and streptococci in logistic regression models and assess non-linearity of this affect using generalised additive models. We calculate NPV for TTP beyond the median value.
The odds ratio for endocarditis from S. aureus bacteria was 0.92 (95%CI 0.85-0.99, p = 0.02) for each hour increase in TTP, with a negative predictive value of TTP greater than the median being 90% (95% CI 85-95%). For streptococci the OR was 1.02 (95%CI 0.98-1.06, p:0.31) and enterococci 1.08 (95% CI 0.99-1.18, p = 0.1) for every increase in hour of TTP.
Longer TTP is associated with lower risk of endocarditis in S. aureus bacteraemia, at 8% reduction per hour, giving an NPV of 90% beyond 13 hours. There was no evidence of association with enterococci or streptococci.
This study looked at whether the time it takes for bacteria to grow in blood (called "time to positivity" or TTP) from patients that are generally unwell, can help predict the risk of heart valve infection (endocarditis) as opposed to other infections. We focused on three common types of bacteria: Staphylococcus aureus, Enterococci, and Streptococci. International guidelines, and standard practice usually recommend heart scans (echocardiography) for everyone with S. aureus infections, even if they don’t have other risk factors, but this is not the case for the other two bacteria. Endocarditis treatment and investigation, has a high burden on patients, with frequent, high doses of antibiotics and invasive tests.
We studied patient records from a large hospital in England to see if TTP could help identify who is at low risk for endocarditis. We found that in patients with S. aureus, a longer TTP was linked to a lower chance of endocarditis. Specifically, for every extra hour before the blood culture turned positive, the risk of endocarditis dropped by about 8%. If the TTP was more than 13 hours, there was a 90% chance that the patient did not have endocarditis. However, for Enterococci and Streptococci, TTP did not help predict the risk.
This information could be helpful with determining who should be prioritised for Echocardiography with S. aures but not the other organisms.
Bacteremia, Endocarditis, Diagnostic stewardship
Staphylococcus aureus bacteremia (SAB) is a common and important cause of infective endocarditis (IE). Currently, the IDSA guidelines for SAB recommend echocardiography for all patients; however, it has been proposed that time-to-positivity (TTP) could be used to predict and stratify patients more likely to have endovascular infections such as IE1,2. One publication of 1703 patient episodes showed that IE reduced by 11% per hour increase in TTP and gave a negative predictive value of 96% at times beyond 13 hours3. Indeed, TTP, in addition to the clinical decision tool VIRSTA, improves the negative predictive value to 100%4. This suggests that obtaining echocardiography for all patients with SAB with a blood culture time to positivity over 13 hours without clinical evidence or specific risk factors for endocarditis may not be necessary.
Other organisms associated with IE include E faecalis (13.6%)5 and some species of high-risk streptococci6, but no clear clinically useful association has been reported between IE and TTP in these organisms7,8, with one study giving an ROC AUC of 0.75 for TTP on IE diagnosis and an association with mortality rates9. TTP is dependent on the generation time of the organism, which is comparable across these causes10–12; and inoculum size, which should be consistent across the same disease process. It is reasonable to explore whether TTP can discriminate between those with and without. These organisms are common causes of bacteremia; however, currently, there is no recommendation to perform echocardiography in all patients, and the European Society of Cardiology 2023 Endocarditis guidance highlights the potential utility of clinical decision tools for the organisms13. With IE prevalence as high as S. aureus, the decision to pursue IE investigations is based on clinical suspicion.
To investigate whether these results are generalizable to cohorts in the UK and to organisms other than S. aureus, we retrospectively investigated our cohort of patients with bacteremia caused by S. aureus, high-risk streptococci and enterococci.
No patient or member of the public was involved in this work
Locally, four quality improvement projects have gathered data on the risk of infective endocarditis (IE) by analyzing isolates from three bacterial genera in recent years: staphylococci (2017–2018 and 2022–2023), enterococci (2020–2023), and streptococci (2017–2024). In this report, we will refer to these by their genus names, which include the following species:
Staphylococci: aureus
Enterococci: faecalis
Streptococci: mitis, oralis, gallolyticus, sanguinis, mutans, gordonii, and cristatus.
Data were extracted from the laboratory systems of a four-hospital network in the UK that serves 2600 inpatient beds, including tertiary cardiac and cardiothoracic patients. Blood cultures were drawn and processed in line with UK SMI S1214 and placed in automated incubators (BD BACTEC FX).
These projects collected data on blood culture results, patient comorbidities, imaging findings, treatment duration, and compliance with relevant guidelines. From these datasets, we extracted the blood culture results, time to positivity, and final diagnoses. Only the first positive blood culture in a patient in a thirty-day period was included. The diagnosis of endocarditis was made by a qualified infection specialist involved in the care of the patient.
Descriptive data of organisms were reported, including positivity rate, mean and median time-to-positivity, and number of cases. Bacteremia was excluded if the time to positivity data was missing or species identification was unavailable. Mann-Whitney tests were performed on each species for IE- and non-IE-associated bacteremia. We explored the association between TTP and IE risk in a) a logistic regression model, calculating the increased risk for each hour of growth, b) a logistic regression model, binarizing TTP at the median TTP, and c) fitting a non-linear model (GAM) to explore inflection points and potential non-linearity. The predictive value of the median TTP was calculated. As transport time to the laboratory can influence time to positivity, we used our cohorts’ median TTP rather than median values reported in the literature15.
Eight hundred and fifty-six bacteraemias were included, of which 46 were removed due to missing TTP data. There were 159 cases of IE: 37 staphylococci (16.2% of 228 bacteremias), 20 enterococci (18.3% of 109), and 97 streptococci (20.5% of 473). The mean TTP was: staphylococci: 14.1 hours (median 13, IQR 10-16); enterococci 11.6 hours (median 11, IQR 9-14); streptococci 14.5 hours (median 13, IQR 10-17). The streptococcal group included cristatus 3, gallolyticus 48, gordonii 13, mitis/oralis 75, mutans 7, sanguinis 34.
The univariate logistic models (IE ~ TTP) produced Odds Ratios of: staphylococci 0.92 (95%CI 0.85-0.99, p:0.02) enterococci 1.08 (95%CI 0.99-1.18, p:0.1) and streptococci 1.02 (95%CI 0.98-1.06, p:0.31) for every increase in hour of TTP. Using growth below the median TTP as a binomial (cutoff) variable produced the OR: staphylococci 2.62 (95% CI 1.25-5.46, p = 0.01), enterococci 0.94 (95% CI 0.36-2.44, p:0.90) and streptococci 0.80 (95% CI 0.52-1.24, p:0.32) (Figure 1).
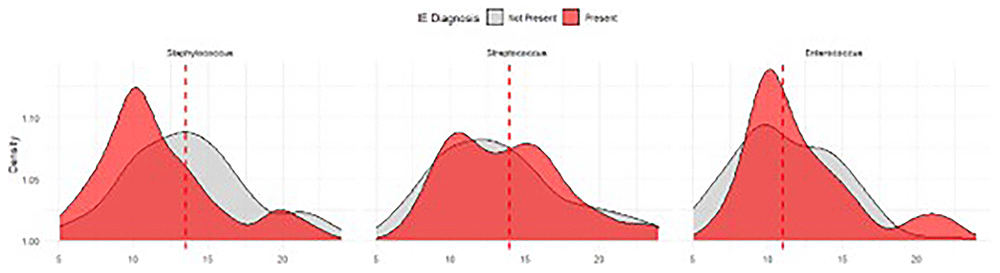
The red dashed lines indicate the species-specific median TTP.
The negative predictive values for growth beyond the median TTP for IE is staphylococci 90% (95% CI 84-95), enterococci 80% (95%CI 68-90) and streptococci 78% (95% CI 73-83). The Generalized additive models exhibited monotonicity and no nonlinearity. These results are shown in the supplementary material.
Our main findings demonstrate that S. aurues bacteremia with longer TTP is significantly less likely to be associated with IE (8% per hour), but this was not true for enterococci or streptococci. Here, S. aureus taking > 13 h to grow had a Negative Predictive Value of 90% (95% CI 85-95). Our study matches the values previously reported by Stromdahl et al., despite the inability to analyze confounding factors in our cohort. This finding highlights the strength of this association. It will be reassuring to infection specialists that patients who cannot tolerate transesophageal echocardiography or are unable to obtain timely imaging can still be risk assessed for endocarditis prior to completing the 2023 ISCVID Duke scoring.
The lack of a relationship between time-to-positivity for streptococci and enterococci may reflect the differences in IE clinical syndromes with these organisms, typically a subacute presentation as opposed to an acute presentation with S. aureus. This assumes that TTP is related to the bacterial density of the original inoculum, which could be due to lower circulating CFU or sample volume16. Notably, the risk of IE remains above 15% across all TTP values for these groups, and there is no clear relationship between the two. While the species-specific analysis of streptococci suggests that S. sanguinis and S. gordonii might have a statistical difference in TTP per IE status, there is a clear overlap in the groups, and clinical significance is not met (Figure 2).
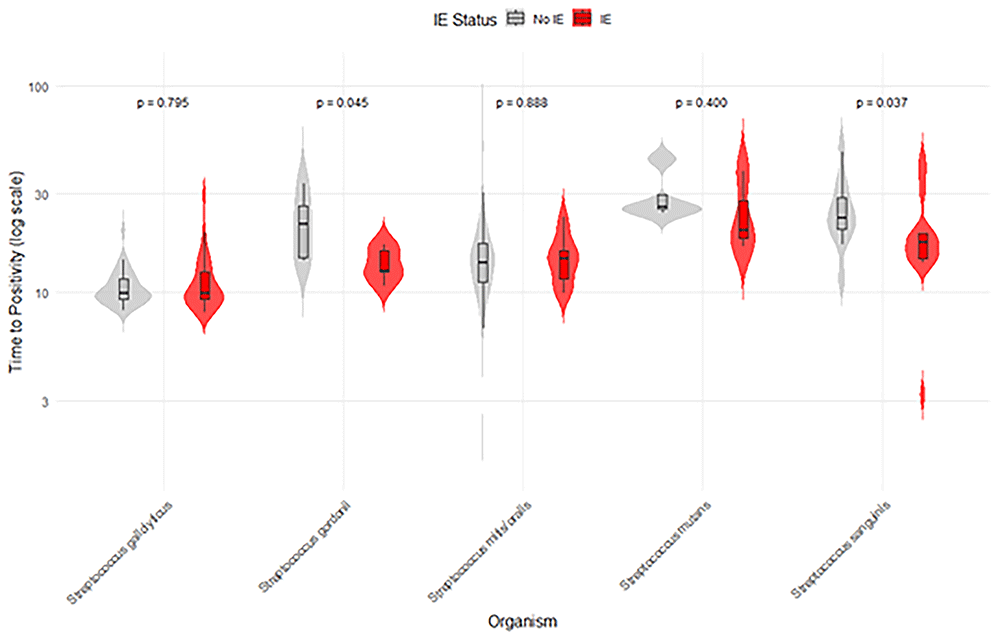
S cristatus removed due to low numbers of positive cases.
We were not able to control for antibiotic exposure prior to sampling, and we were only able to duplicate the culture results from within the hospital network. IE was diagnosed by an infection specialist in all cases, but 2023 ISCVID Duke's criteria were not extracted in this study.
A longer TTP is associated with lower rates of IE in S. aureus bacteremia at 8% reduction per hour, giving an NPV of 90% beyond 13 hours. There was no evidence of an association with enterococci or streptococci.
Four reviews of bacteremias were undertaken as part of quality improvement projects and granted local permission to collect patient identifiable data for these purposes (North Bristol Trust, and University Hospitals Bristol and Weston). Data taken from these reviews included only the TTP results and IE diagnosis. Therefore, formal ethics approval was not required.
Data is available as deidentified participant data and analysis code. Request should be made to the lead author at gavin.deas@nbt.nhs.uk. Local IRB approval will be required for any data sharing. Data will be available to any researcher whose proposed use of the data has been approved with investigator support and fully executed data use agreement. Supplementary material: Deas, G. (2025, August 9). Time to positivity and endocarditis. Retrieved from osf.io/w5cqg
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Register with NIHR Open Research
Already registered? Sign in
If you are a previous or current NIHR award holder, sign up for information about developments, publishing and publications from NIHR Open Research.
We'll keep you updated on any major new updates to NIHR Open Research
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)